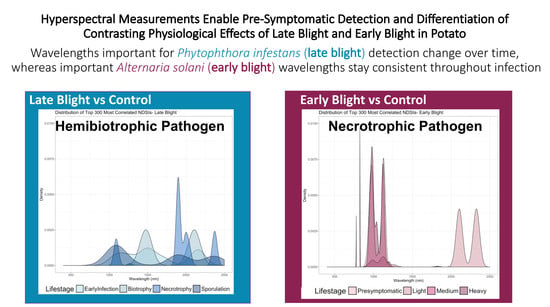
Hyperspectral Measurements Enable Pre-Symptomatic Detection and Differentiation of Contrasting Physiological Effects of Late Blight and Early Blight in Potato
Abstract
:1. Introduction
2. Materials & Methods
2.1. Plant & Pathogen Materials
2.2. Experimental Set-Up
2.3. Clearing and Staining
2.4. Reflectance Measurements
2.5. Data Preparation
2.6. Data Analysis
3. Results
3.1. Phytophthora infestans Life Cycle Can Be Accurately Characterized with our Disease Time Rating Scale
3.2. Disease Effects of Phytophthora infestans and Alternaria solani Can Be Detected and Differentiated across All Time Points Using Spectroscopy
3.3. Plant Response to P. Infestans, A. solani, and Co-Inoculations Can Be Differentiated from Healthy Leaves Prior to Symptom Development with Spectroscopy
3.4. Spectral Profiles of Infected Leaves Corroborate Pathogen Biology
4. Discussion
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- FAO—News Article: New Standards to Curb the Global Spread of Plant Pests and Diseases. Available online: http://www.fao.org/news/story/en/item/1187738/icode/ (accessed on 20 November 2019).
- Mahlein, A.-K.; Kuska, M.T.; Behmann, J.; Polder, G.; Walter, A. Hyperspectral Sensors and Imaging Technologies in Phytopathology: State of the Art. Annu. Rev. Phytopathol. 2018, 56, 535–558. [Google Scholar] [CrossRef] [PubMed]
- Curran, P.J. Remote sensing of foliar chemistry. Remote Sens. Environ. 1989, 30, 271–278. [Google Scholar] [CrossRef]
- Kokaly, R.F.; Asner, G.P.; Ollinger, S.V.; Martin, M.E.; Wessman, C.A. Characterizing canopy biochemistry from imaging spectroscopy and its application to ecosystem studies. Remote Sens. Environ. 2009, 113, S78–S91. [Google Scholar] [CrossRef]
- Kokaly, R.F.; Skidmore, A.K. Plant phenolics and absorption features in vegetation reflectance spectra near 1.66 μm. Int. J. Appl. Earth Obs. Geoinf. 2015, 43, 55–83. [Google Scholar] [CrossRef] [Green Version]
- Serbin, S.P. Spectroscopic Determination of Leaf Nutritional, Morpholgical, and Metabolic Traits. Ph.D. Thesis, University of Wisconsin-Madison, Madison, WI, USA, May 2012. [Google Scholar]
- Serbin, S.P.; Singh, A.; McNeil, B.E.; Kingdon, C.C.; Townsend, P.A. Spectroscopic determination of leaf morphological and biochemical traits for northern temperate and boreal tree species. Ecol. Appl. 2014, 24, 1651–1669. [Google Scholar] [CrossRef] [Green Version]
- Couture, J.J.; Serbin, S.P.; Townsend, P.A. Spectroscopic sensitivity of real-time, rapidly induced phytochemical change in response to damage. New Phytol. 2013, 198, 311–319. [Google Scholar] [CrossRef]
- Couture, J.J.; Singh, A.; Rubert-Nason, K.F.; Serbin, S.P.; Lindroth, R.L.; Townsend, P.A. Spectroscopic determination of ecologically relevant plant secondary metabolites. Methods Ecol. Evol. 2016, 7, 1402–1412. [Google Scholar] [CrossRef] [Green Version]
- Singh, A.; Serbin, S.P.; McNeil, B.E.; Kingdon, C.C.; Townsend, P.A. Imaging spectroscopy algorithms for mapping canopy foliar chemical and morphological traits and their uncertainties. Ecol. Appl. 2015, 25, 2180–2197. [Google Scholar] [CrossRef]
- Asner, G.P.; Martin, R.E. Spectral and chemical analysis of tropical forests: Scaling from leaf to canopy levels. Remote Sens. Environ. 2008, 112, 3958–3970. [Google Scholar] [CrossRef]
- Asner, G.P.; Martin, R.E.; Anderson, C.B.; Knapp, D.E. Quantifying forest canopy traits: Imaging spectroscopy versus field survey. Remote Sens. Environ. 2015, 158, 15–27. [Google Scholar] [CrossRef]
- Cavender-Bares, J.; Meireles, J.E.; Couture, J.J.; Kaproth, M.A.; Kingdon, C.C.; Singh, A.; Serbin, S.P.; Center, A.; Zuniga, E.; Pilz, G.; et al. Associations of Leaf Spectra with Genetic and Phylogenetic Variation in Oaks: Prospects for Remote Detection of Biodiversity. Remote Sens. 2016, 8, 221. [Google Scholar] [CrossRef] [Green Version]
- Yuan, M.; Couture, J.J.; Townsend, P.A.; Ruark, M.D.; Bland, W.L. Spectroscopic Determination of Leaf Nitrogen Concentration and Mass Per Area in Sweet Corn and Snap Bean. Agron. J. 2016, 108, 2519–2526. [Google Scholar] [CrossRef]
- Carter, G.A.; Knapp, A.K. Leaf optical properties in higher plants: Linking spectral characteristics to stress and chlorophyll concentration. Am. J. Bot. 2001, 88, 677–684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agrios, G. Plant Pathogens and Disease: General Introduction; Elsevier: Amsterdam, The Netherland, 2009. [Google Scholar]
- Mahlein, A.-K. Plant Disease Detection by Imaging Sensors—Parallels and Specific Demands for Precision Agriculture and Plant Phenotyping. Plant Dis. 2015, 100, 241–251. [Google Scholar] [CrossRef] [Green Version]
- Arens, N.; Backhaus, A.; Döll, S.; Fischer, S.; Seiffert, U.; Mock, H.-P. Non-invasive Presymptomatic Detection of Cercospora beticola Infection and Identification of Early Metabolic Responses in Sugar Beet. Front. Plant Sci. 2016, 7, 1377. [Google Scholar] [CrossRef] [Green Version]
- Couture, J.J.; Singh, A.; Charkowski, A.O.; Groves, R.L.; Gray, S.M.; Bethke, P.C.; Townsend, P.A. Integrating Spectroscopy with Potato Disease Management. Plant Dis. 2018, 102, 2233–2240. [Google Scholar] [CrossRef] [Green Version]
- Heim, R.H.J.; Wright, I.J.; Chang, H.-C.; Carnegie, A.J.; Pegg, G.S.; Lancaster, E.K.; Falster, D.S.; Oldeland, J. Detecting myrtle rust (Austropuccinia psidii) on lemon myrtle trees using spectral signatures and machine learning. Plant Pathol. 2018, 67, 1114–1121. [Google Scholar] [CrossRef]
- Gold, K.M.; Gevens, A.J.; Townsend, P.A. System for Detection of Disease in Plants. U.S. Patent App. 16/251,415, 1 June 2019. [Google Scholar]
- Gold, K.M.; Townsend, P.A.; Herrmann, I.; Gevens, A.J. Investigating potato late blight physiological differences across potato cultivars with spectroscopy and machine learning. Plant Sci. 2019, 110316. [Google Scholar] [CrossRef]
- Oerke, E.-C.; Leucker, M.; Steiner, U. Sensory assessment of Cercospora beticola sporulation for phenotyping the partial disease resistance of sugar beet genotypes. Plant Methods 2019, 15, 133. [Google Scholar] [CrossRef] [Green Version]
- Fry, W.E. The Canon of Potato Science: 10. Late Blight and Early Blight. Potato Res. 2007, 50, 243–245. [Google Scholar] [CrossRef]
- Fry, W.E.; Birch, P.R.J.; Judelson, H.S.; Grünwald, N.J.; Danies, G.; Everts, K.L.; Gevens, A.J.; Gugino, B.K.; Johnson, D.A.; Johnson, S.B.; et al. Five Reasons to Consider Phytophthora infestans a Reemerging Pathogen. Phytopathology 2015, 105, 966–981. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haverkort, A.J.; Boonekamp, P.M.; Hutten, R.; Jacobsen, E.; Lotz, L.A.P.; Kessel, G.J.T.; Visser, R.G.F.; Van Der Vossen, E.A.G. Societal costs of late blight in potato and prospects of durable resistance through cisgenic modification. Potato Res. 2008, 51, 47–57. [Google Scholar] [CrossRef]
- Fry, W. Phytophthora infestans: The plant (and R gene) destroyer. Mol. Plant Pathol. 2008, 9, 385–402. [Google Scholar] [CrossRef] [PubMed]
- Pasche, J.S.; Piche, L.M.; Gudmestad, N.C. Effect of the F129L Mutation in Alternaria solani on Fungicides Affecting Mitochondrial Respiration. Plant Dis. 2005, 89, 269–278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leiminger, J.H.; Adolf, B.; Hausladen, H. Occurrence of the F129L mutation in Alternaria solani populations in Germany in response to QoI application, and its effect on sensitivity. Plant Pathol. 2014, 63, 640–650. [Google Scholar] [CrossRef]
- Nishimura, S.; Kohmoto, K. Host-Specific Toxins and Chemical Structures from Alternaria Species. Annu. Rev. Phytopathol. 1983, 21, 87–116. [Google Scholar] [CrossRef]
- Wu, D.; Feng, L.; Zhang, C.; He, Y. Early Detection of Botrytis cinerea on Eggplant Leaves Based on Visible and Near-Infrared Spectroscopy. Trans. ASABE 2008, 51, 1133–1139. [Google Scholar] [CrossRef]
- Rumpf, T.; Mahlein, A.-K.; Steiner, U.; Oerke, E.-C.; Dehne, H.-W.; Plümer, L. Early detection and classification of plant diseases with Support Vector Machines based on hyperspectral reflectance. Comput. Electron. Agric. 2010, 74, 91–99. [Google Scholar] [CrossRef]
- Kuska, M.; Wahabzada, M.; Leucker, M.; Dehne, H.-W.; Kersting, K.; Oerke, E.-C.; Steiner, U.; Mahlein, A.-K. Hyperspectral phenotyping on the microscopic scale: Towards automated characterization of plant-pathogen interactions. Plant Methods 2015, 11, 28. [Google Scholar] [CrossRef] [Green Version]
- Xie, C.; Yang, C.; He, Y. Hyperspectral imaging for classification of healthy and gray mold diseased tomato leaves with different infection severities. Comput. Electron. Agric. 2017, 135, 154–162. [Google Scholar] [CrossRef]
- Zarco-Tejada, P.J.; Camino, C.; Beck, P.S.A.; Calderon, R.; Hornero, A.; Hernández-Clemente, R.; Kattenborn, T.; Montes-Borrego, M.; Susca, L.; Morelli, M.; et al. Previsual symptoms of Xylella fastidiosa infection revealed in spectral plant-trait alterations. Nat. Plants 2018, 4, 432–439. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Qin, Z.; Liu, X. Remote Sensed Spectral Imagery to Detect Late Blight in Field Tomatoes. Precis. Agric. 2005, 6, 489–508. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, M.; Zhu, J.; Geng, S. Spectral prediction of Phytophthora infestans infection on tomatoes using artificial neural network (ANN). Int. J. Remote Sens. 2008, 29, 1693–1706. [Google Scholar] [CrossRef]
- Baranowski, P.; Jedryczka, M.; Mazurek, W.; Babula-Skowronska, D.; Siedliska, A.; Kaczmarek, J. Hyperspectral and Thermal Imaging of Oilseed Rape (Brassica napus) Response to Fungal Species of the Genus Alternaria. PLoS ONE 2015, 10, e0122913. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, C.; Shao, Y.; Li, X.; He, Y. Detection of early blight and late blight diseases on tomato leaves using hyperspectral imaging. Sci. Rep. 2015, 5, 16564. [Google Scholar] [CrossRef]
- Sugiura, R.; Tsuda, S.; Tamiya, S.; Itoh, A.; Nishiwaki, K.; Murakami, N.; Shibuya, Y.; Hirafuji, M.; Nuske, S. Field phenotyping system for the assessment of potato late blight resistance using RGB imagery from an unmanned aerial vehicle. Biosyst. Eng. 2016, 148, 1–10. [Google Scholar] [CrossRef]
- Duarte-Carvajalino, J.M.; Alzate, D.F.; Ramirez, A.A.; Santa-Sepulveda, J.D.; Fajardo-Rojas, A.E.; Soto-Suárez, M. Evaluating Late Blight Severity in Potato Crops Using Unmanned Aerial Vehicles and Machine Learning Algorithms. Remote Sens. 2018, 10, 1513. [Google Scholar] [CrossRef] [Green Version]
- Bienkowski, D.; Aitkenhead, M.J.; Lees, A.K.; Gallagher, C.; Neilson, R. Detection and differentiation between potato (Solanum tuberosum) diseases using calibration models trained with non-imaging spectrometry data. Comput. Electron. Agric. 2019, 167, 105056. [Google Scholar] [CrossRef]
- Franceschini, M.H.D.; Bartholomeus, H.; van Apeldoorn, D.F.; Suomalainen, J.; Kooistra, L. Feasibility of Unmanned Aerial Vehicle Optical Imagery for Early Detection and Severity Assessment of Late Blight in Potato. Remote Sens. 2019, 11, 224. [Google Scholar] [CrossRef] [Green Version]
- Lamour, K.H.; Hausbeck, M.K. The Spatiotemporal Genetic Structure of Phytophthora capsici in Michigan and Implications for Disease Management. Phytopathology 2002, 92, 681–684. [Google Scholar] [CrossRef] [Green Version]
- Fry, W.E.; Patev, S.P.; Myers, K.L.; Bao, K.; Fei, Z. Phytophthora infestans Sporangia Produced in Culture and on Tomato Leaflet Lesions Show Marked Differences in Indirect Germination Rates, Aggressiveness, and Global Transcription Profiles. Mol. Plant Microbe Interact. 2018, 32, 515–526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barratt, R.; Horsfall, J. An improved grading system for measuring plant disease. Phytopathology 1945, 35, 655. [Google Scholar]
- Gold, K.M.; Townsend, P.A.; Larson, E.R.; Herrmann, I.; Gevens, A.J. Contact reflectance spectroscopy for rapid, accurate, and non-destructive Phytophthora infestans clonal lineage discrimination. Phytopathology 2019. Available online: https://apsjournals.apsnet.org/doi/abs/10.1094/PHYTO-08-19-0294-R (accessed on 7 January 2020). [CrossRef] [PubMed] [Green Version]
- Yin, J.; Gu, B.; Huang, G.; Tian, Y.; Quan, J.; Lindqvist-Kreuze, H.; Shan, W. Conserved RXLR Effector Genes of Phytophthora infestans Expressed at the Early Stage of Potato Infection Are Suppressive to Host Defense. Front. Plant Sci. 2017, 81, 2155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Villani, T.S.; Koroch, A.R.; Simon, J.E. An improved clearing and mounting solution to replace chloral hydrate in microscopic applications. Appl. Plant Sci. 2013, 1, 1300016. [Google Scholar] [CrossRef]
- Herrmann, I.; Vosberg, S.K.; Ravindran, P.; Singh, A.; Chang, H.-X.; Chilvers, M.I.; Conley, S.P.; Townsend, P.A. Leaf and Canopy Level Detection of Fusarium Virguliforme (Sudden Death Syndrome) in Soybean. Remote Sens. 2018, 10, 426. [Google Scholar] [CrossRef] [Green Version]
- Zhai, Y.; Cui, L.; Zhou, X.; Gao, Y.; Fei, T.; Gao, W. Estimation of nitrogen, phosphorus, and potassium contents in the leaves of different plants using laboratory-based visible and near-infrared reflectance spectroscopy: Comparison of partial least-square regression and support vector machine regression methods. Int. J. Remote Sens. 2013, 34, 2502–2518. [Google Scholar]
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’hara, R.; Simpson, G.L.; Solymos, P.; Stevens, M.H.H.; Wagner, H.; et al. Package ‘vegan.’. Community Ecol. Package Version 2013, 2, 1–295. [Google Scholar]
- Mevik, B.-H.; Wehrens, R.; Liland, K.H. pls: Partial least squares and principal component regression. R Package Version 2011, 2, 3. [Google Scholar]
- Kuhn, M. caret: Classification and Regression Training. Astrophys. Source Code Libr. 2015. [Google Scholar]
- Worley, B.; Powers, R. Multivariate analysis in metabolomics. Curr. Metab. 2013, 1, 92–107. [Google Scholar]
- Wright, I.J.; Reich, P.B.; Westoby, M.; Ackerly, D.D.; Baruch, Z.; Bongers, F.; Cavender-Bares, J.; Chapin, T.; Cornelissen, J.H.C.; Diemer, M.; et al. The worldwide leaf economics spectrum. Nature 2004, 428, 821. [Google Scholar] [CrossRef] [PubMed]
- Reich, P.B. The world-wide ‘fast–slow’ plant economics spectrum: A traits manifesto. J. Ecol. 2014, 102, 275–301. [Google Scholar] [CrossRef]
- Gao, B. NDWI—A normalized difference water index for remote sensing of vegetation liquid water from space. Remote Sens. Environ. 1996, 58, 257–266. [Google Scholar] [CrossRef]
- Pinheiro, J.; Bates, D.; DebRoy, S.; Sarkar, D.; R Core Team. nlme: Linear and nonlinear mixed effects models. R Package Version 3.1–111. Comput. Man. 2013. [Google Scholar]
- Lenth, R. Emmeans: Estimated marginal means, aka least-squares means. R Package Vers. 2018, 1, 2. [Google Scholar]
- Jackson, R.D. Remote Sensing of Biotic and Abiotic Plant Stress. Annu. Rev. Phytopathol. 1986, 24, 265–287. [Google Scholar] [CrossRef]
- Nilsson, H. Remote Sensing and Image Analysis in Plant Pathology. Annu. Rev. Phytopathol. 1995, 33, 489–528. [Google Scholar] [CrossRef]
- Hatfield, P.L.; Pinter, P.J. Remote sensing for crop protection. Crop Prot. 1993, 12, 403–413. [Google Scholar] [CrossRef]
- Zuluaga, A.P.; Vega-Arreguín, J.J.C.; Fei, Z.; Ponnala, L.; Lee, S.J.; Matas, A.J.; Patev, S.; Fry, W.E.; Rose, J.K.C. Transcriptional dynamics of Phytophthora infestans during sequential stages of hemibiotrophic infection of tomato. Mol. Plant Pathol. 2016, 17, 29–41. [Google Scholar] [CrossRef]
- Henriquez, M.A.; Adam, L.R.; Daayf, F. Alteration of secondary metabolites’ profiles in potato leaves in response to weakly and highly aggressive isolates of Phytophthora infestans. Plant Physiol. Biochem. 2012, 57, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Townsend, P.A.; Schweiger, A.K.; Couture, J.J.; Singh, A.; Hobbie, S.E.; Cavender-Bares, J. Mapping foliar functional traits and their uncertainties across three years in a grassland experiment. Remote Sens. Environ. 2019, 221, 405–416. [Google Scholar] [CrossRef]
- Galeano Garcia, P.; Neves dos Santos, F.; Zanotta, S.; Eberlin, M.N.; Carazzone, C. Metabolomics of Solanum lycopersicum Infected with Phytophthora infestans Leads to Early Detection of Late Blight in Asymptomatic Plants. Molecules 2018, 23, 3330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Erwin, D.C.; Ribeiro, O.K. Phytophthora Diseases Worldwide; APS Press: St. Paul, MN, USA, 1996. [Google Scholar]





| Late Blight | ||||||
|---|---|---|---|---|---|---|
| Kappa (val) | Model | |||||
| All Timepoints | Early Infection | Biotrophy | Necrotrophy | Sporulation | ||
| Data | All Timepoints | 0.766 | 0.608 | 0.203 | 0.454 | 0.535 |
| Early Infection | 0.857 | 0.872 | 0.207 | 0.476 | 0.441 | |
| Biotrophy | 0.753 | 0.559 | 0.812 | 0.286 | 0.410 | |
| Necrotrophy | 0.706 | 0.424 | 0.180 | 0.790 | 0.731 | |
| Sporulation | 0.722 | 0.375 | 0.044 | 0.596 | 0.636 | |
| Early Blight | |||||
|---|---|---|---|---|---|
| Kappa (val) | Model | ||||
| All Timepoints | Pre-Symptomatic | Light | Heavy | ||
| Data | All Timepoints | 0.904 | 0.709 | 0.626 | 0.623 |
| Pre-Symptomatic | 0.935 | 0.936 | 0.878 | 0.844 | |
| Light | 0.984 | 0.957 | 0.984 | 0.977 | |
| Heavy | 0.951 | 0.849 | 0.965 | 0.982 | |
| All Timepoints Kappa = 0.83 Components = 14 | Early Blight | Late Blight | Actual # of samples per class | Producer’s Accuracy % | |
| Early Blight | 58.93 | 11.07 | 70 | 84.19 | |
| Late Blight | 1.56 | 82.44 | 84 | 98.14 | |
| Total # of classified samples | 60.49 | 93.51 | Total Accuracy: 91.8% | ||
| User’s Accuracy % | 97.46 | 88.27 | |||
| Pre-Symptomatic Kappa = 0.78 Components = 12 | Early Blight | Late Blight | Actual # of samples per class | Producer’s Accuracy % | |
| Early Blight | 14.47 | 2.53 | 17 | 85.12 | |
| Late Blight | 3.01 | 43.99 | 47 | 93.60 | |
| Total # of classified samples | 17.48 | 46.52 | Total Accuracy: 91.3% | ||
| User’s Accuracy % | 83.65 | 94.68 | |||
| Post-Symptomatic Light Kappa = 0.86 Components = 16 | Early Blight | Late Blight | Actual # of samples per class | Producer’s Accuracy % | |
| Early Blight | 43.17 | 6.83 | 50 | 86.34 | |
| Late Blight | 2.06 | 90.94 | 93 | 97.78 | |
| Total # of classified samples | 45.23 | 97.77 | Total Accuracy: 93.8% | ||
| User’s Accuracy % | 95.51 | 93.07 | |||
| Post-Symptomatic Heavy Kappa = 0.94 Components = 5 | Early Blight | Late Blight | Actual # of samples per class | Producer’s Accuracy % | |
| Early Blight | 15.81 | 1.19 | 17 | 93.00 | |
| Late Blight | 0 | 21 | 21 | 100.00 | |
| Total # of classified samples | 15.81 | 22.19 | Total Accuracy: 96.9% | ||
| User’s Accuracy % | 100.00 | 94.80 | |||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gold, K.M.; Townsend, P.A.; Chlus, A.; Herrmann, I.; Couture, J.J.; Larson, E.R.; Gevens, A.J. Hyperspectral Measurements Enable Pre-Symptomatic Detection and Differentiation of Contrasting Physiological Effects of Late Blight and Early Blight in Potato. Remote Sens. 2020, 12, 286. https://0-doi-org.brum.beds.ac.uk/10.3390/rs12020286
Gold KM, Townsend PA, Chlus A, Herrmann I, Couture JJ, Larson ER, Gevens AJ. Hyperspectral Measurements Enable Pre-Symptomatic Detection and Differentiation of Contrasting Physiological Effects of Late Blight and Early Blight in Potato. Remote Sensing. 2020; 12(2):286. https://0-doi-org.brum.beds.ac.uk/10.3390/rs12020286
Chicago/Turabian StyleGold, Kaitlin M., Philip A. Townsend, Adam Chlus, Ittai Herrmann, John J. Couture, Eric R. Larson, and Amanda J. Gevens. 2020. "Hyperspectral Measurements Enable Pre-Symptomatic Detection and Differentiation of Contrasting Physiological Effects of Late Blight and Early Blight in Potato" Remote Sensing 12, no. 2: 286. https://0-doi-org.brum.beds.ac.uk/10.3390/rs12020286