1. Introduction
Rice (
Oryza sativa L.) is the second most cultivated cereal crop and the most consumed staple food in the world, since more than three billion people rely on rice as their primary source of food. Although it is predominant in Asia, rice has also been cultivated in Europe since the 15th century, mainly in Mediterranean countries including Italy, Spain, Portugal, Greece, and France (FAO database 2018). Rice is cultivated under a wide range of ecosystems, but more than 90% of the world’s rice production is harvested from irrigated or rainfed lowland rice fields [
1]. Thus, increase in rice production is needed if the increased demand from population growth is to be met.
The use of nitrogen (N) fertilizer has changed the global N cycle markedly and has been causing various negative environmental consequences, such as eutrophication of surface water, global warming, and ozone layer depletion [
2,
3,
4]. Modern production agriculture requires efficient, sustainable, and environmentally-sound management practices. N is a key factor in achieving optimum lowland rice grain yields [
5]. It is the nutrient input normally required in large quantities for achieving high yields, but soils under these conditions are saturated, flooded, and anaerobic and, therefore, N use efficiency is low [
6]. However, more than 50% of the applied N is not assimilated by the rice plant and it is lost through different mechanisms including ammonia volatilization, surface runoff, nitrification-denitrification, and leaching [
7,
8,
9]. N
2O is the main greenhouse gas (GHG) related to agricultural soil emissions, essentially due to microbial transformation of nitrogen in the soil. This concerns N mineral fertilizers, manure spreading, and N from crop residues incorporated into the soil or lixiviation of surplus nitrogen. N
2O has high global warming potential (298 times higher than CO
2) and it should be minimized to reduce agricultural GHG emissions in total. The application of mineral N in the form of chemical fertilizers can also increase the N
2O emissions [
10]. A recent study estimated the seasonal direct emission of N
2O from the paddy rice system in China (a country accounting for approximately 30% of the global rice production) to be 31.1 Gg N
2O-N for 2014, analyzing data from multiple studies [
11]. Another recent study reported that intermittent flooding practices in rice can significantly increase N
2O emissions [
12], although there is an active debate on this issue [
13,
14]. Therefore, N fertilizer management strategies that increase crop productivity and N use efficiency, while reducing negative environmental consequences, have to focus on parameters such as optimum time, rate, and spatial distribution methods that synchronize plant N requirements with N supply, in order to reduce N losses and maximize uptake of applied N in the crop [
15]. Legislative measures adopted to comply with the Directive 91/676/EEC concerning the “Protection of Waters against Pollution caused by Nitrates from Agricultural Sources,” in some cases did not obtain the expected results and are not always accepted (or complied with) by farmers [
16,
17].
A key contributor to yield increases will be the efficient and effective use of nitrogen (N) fertilizer, which is relatively low in irrigated rice, because of rapid N losses from volatilization and denitrification in the soil-floodwater system [
18]. The “4R” nutrient stewardship—applying the right nutrient source at the right rate, at the right time, and in the right place—is an innovative approach in fertilizer management [
19]. Precision agriculture also has a positive impact on farm productivity and economics, as it provides higher or equal yields with lower production cost than conventional practices. The adoption of precision agriculture techniques for N management has the potential for improving agronomic, economic, and environmental efficiency in the use of such input [
20].
Regarding the rest of the fertilization practices generally followed in Europe, phosphorous and potassium are supplied in the pre-planting stage at 50–70 kg∙ha
−1 and 100–150 kg∙ha
−1, respectively. The first fertilization intervention usually provides a nitrogen–phosphorous–potassium complex; it is carried out before the field flooding. The second supply is usually applied when rice plants are at the 3–5-leaf stage, at the beginning of tillering. A third fertilization can sometimes occur during panicle initiation. Sulfur can be applied using sulfuric fertilizers (NH
4 or K) and zinc is generally needed in soils with high pH, whereas calcium is needed in pathogenic soils with high salinity. The latter elements are used in Europe occasionally or after deficiencies [
21].
A major challenge in N management through precision agriculture techniques is the accurate prediction and mapping of plant agronomic traits related to plant nutrient status and—most importantly—to N content. The traditional soil-based testing methods widely used for upland crops are not suitable for N recommendations in rice fields due to the flooding and complexity of N cycling in the rice paddy’s soil during the growing period [
22]. Even before flooding, the available soil-N test is not accurate enough to lead to confident recommendations for N fertilizer applications [
23]. Remote sensing technologies offer a viable alternative for deriving precise N recommendations in rice fields through the dynamic non-destructive estimation of plant N status throughout the growing season, along with predictions of other agronomic traits of interest (e.g., yield). Coupled with the rapidly advancing technology of unmanned aerial vehicle (UAV) platforms, they are nowadays starting to offer cost-effective solutions to various aspects of crop monitoring and sustainable crop management [
24,
25,
26,
27].
A few studies have proposed employing the traditional approach of active canopy sensors for estimating yield-related agronomic traits of rice plants, which can help in supporting precision N management [
28,
29,
30,
31,
32,
33,
34,
35]. For active field applications, these sensors could be installed on the machinery for real-time sensing, but such systems are not common in rice cultivation [
36]. Most importantly, their use does become cost-inefficient (time and energy consuming) if the tractor must enter the rice paddy just for monitoring the N status, at growth stages other than the appropriate for fertilizer application or for weed and pest management. A substantial number of studies have also investigated the relationship between narrow-band vegetation indices (VIs) and various agronomic traits of rice plants [
22,
37,
38,
39,
40,
41,
42,
43,
44,
45,
46,
47,
48,
49,
50,
51,
52,
53,
54]. The VIs are calculated from the spectral signatures obtained from handheld spectrometers, which are portable non-imaging hyperspectral sensors acquiring single spectral signatures from a small (typically circular) surface over the canopy. Most of these studies construct empirical models through linear regression for each possible pair of agronomic traits versus VIs, thus identifying the individual VIs that are highly correlated with each agronomic trait, but using the whole hyperspectral signatures as input to multivariable N status prediction models has also been proposed [
45,
48].
The results of the aforementioned studies are important, since they have identified the spectral wavelengths and/or VIs that are highly correlated with yield-related traits (e.g., [
32,
33,
46,
52]) or even proposed N recommendation methodologies based on remotely sensed data [
28]. However, canopy-level spectral data from spectrometers becomes inappropriate for fine-scale precision farming, since that would require an extremely time-consuming and cost-inefficient dense sampling. In addition, the calculation of optimized VIs requires the sensor to incorporate bands at very specific wavelengths, which are typically not available in most multispectral sensors. It is true that compact hyperspectral sensors that can be mounted onboard UAVs (or movable structures above the canopy) have been developed the last few years and have been successfully employed for estimating rice agronomic traits [
55,
56,
57]. Yet, the use of these systems is still limited in precision agriculture applications, their cost is relatively high, and processing such large volumes of data is quite challenging, although they have great potential for being operationally employed in the future.
Multispectral imaging sensors probably constitute the most appropriate system for precision agriculture applications in rice cultivation. Traditionally, satellite imagery has been employed for monitoring rice growth and agronomic traits. Medium-resolution satellite imagery (spatial resolution of 20–30 m) has been employed for providing rather coarse estimations of rice crop traits [
58,
59,
60,
61] or has been incorporated within rice growth simulation models [
62]. High-resolution synthetic aperture radar (SAR) data have been employed for estimating morphological traits (most notably height) [
63]. Recently, Sentinel-2 data have also been employed for estimating leaf area index (LAI) and plant N concentration of rice fields through empirical regression modeling, which were then used for producing N nutritional index maps [
64]. Finally, a few studies have used commercial high-resolution (spatial resolution less than 10 m) satellite imagery to estimate N status [
36,
65,
66] or within-field variability [
61]. However, the most important drawback of high-resolution satellite images is their high cost for real-life applications, as almost all vendors enforce a large minimum area that can be ordered for a single image. If multiple images within the season are required for providing N recommendations and taking into consideration that rice paddies in a single cultivation have variable sowing dates—typically up to 30 days difference (due to genotypic differences in plant stages etc.)—the cost increases exponentially.
With the rapid development of UAV platforms (load capacity and flying autonomy), compact and lightweight multispectral sensors onboard UAVs seems to be the most direct and cost-efficient approach to precision agriculture in rice today. However, very few studies have been published that exploit such systems for estimating agronomic traits of rice plants. Initially, plain RGB cameras were used for estimating N status of rice plants [
67,
68], but these cameras are difficult to calibrate for different lighting conditions and they do not incorporate a near-infrared (NIR) channel that is required for the calculation of most VIs. Modifying the camera with a polarizing filter could solve the latter problem [
69], but the sensor’s spectral response is still not appropriate for accurate calculation of most VIs, which require specific ranges of wavelengths. Good correlations of specific VIs calculated from multispectral sensors (RGB plus NIR channel) and measurements from chlorophyll meters (SPAD meter) have been reported [
70,
71]. However, estimating leaf N content from SPAD readings is not straightforward, since significant variability has been observed between SPAD values and both leaf chlorophyll [
72,
73,
74] and leaf N [
75,
76,
77] content. Finally, Lu et al. [
78] has tested the potential of a 5-band multispectral sensor (RGB plus red-edge (RE) and NIR channels) onboard a UAV for estimating LAI and N content of rice plants, through simple linear regression. Medium to high correlations were reported at the panicle growing stage—close to the second top-dressing fertilization (TdF)—with lower coefficient of determination (R
2) values reported for the stem elongation stage (close to the first TdF). However, their experimentation considered field data from a single growing season, and the lower R
2 values reported for the stem elongation stage suggest that multiple remote sensing variables must be considered simultaneously for increasing the modeling accuracy.
The objective of the current paper is to present empirical models for predicting agronomic traits of rice plants through remotely-sensed multispectral imagery. Models for predicting plant height, aboveground biomass, N concentration, Ν uptake, grain yield, and harvest index have been constructed following a multiple linear regression approach at different growth stages (at tillering and at booting), using as input reflectance values and VIs obtained from a compact four-band sensor (green, red, narrow-band RE, and NIR channels) onboard a UAV. The models were constructed using field data and images from two consecutive years in a number of experimental rice plots in Greece, employing different N treatments. Models for both estimating the current crop status (e.g., N uptake at the time of image acquisition) and predicting the future one (e.g., N uptake of grains at maturity) were developed and evaluated. To the best of our knowledge, this is the first study that presents such a comprehensive analysis for estimating various yield-related agronomic traits of rice plants during the growing season from UAV-collected multispectral data.
3. Results
During both rice cultivation periods, temperature and relative humidity were constantly recorded in order to compare the meteorological data sets.
Table 4 reports monthly averages of temperature and relative humidity during the two growing seasons of the experimentation. The second year (2017) was slightly colder and less humid; however, these differences were minor and it can be concluded that they could not induce alterations in the morphophysiology of the rice plants between the two experiments over the two years.
Figure 2 presents the differences of selected agronomic traits with respect to the fertilization treatment. Biomass, N uptake, and yield exhibit a rather expected trend, that is, higher values with increased amount of N fertilizer applied. Yield also exhibits the expected saturation with N fertilization, that is, the great differences in NU and BT between C2 (standard local practice) and C3 (double amount) at BBCH 45 were not fully reflected in yield differences. The variability was much higher at BBCH 25 (at tillering), since the canopy was not yet fully closed, due to the fact that the tillers were not completely developed and no TdF had been applied yet. As the growing season continued, the differences became more pronounced. The aforementioned trend was not clearly exhibited for N concentration and HI, which was also reflected in the modeling process that resulted in generally low predictive accuracy for those traits, as will be shown in the following. A possible explanation for this behavior is provided in the Discussion section. For completeness,
Table 5 reports the results of a one-way ANOVA statistical analysis, followed by Tukey’s honest significant difference (HSD) post-hoc test [
100]. The results of the latter are compactly presented by means of labeled groups (letters), with two treatments that include the same letter denoting statistically non-significant differences between the mean values of the corresponding agronomic trait (e.g., treatments C1 and C2 for the Yield trait were not found to exhibit statistically significant differences, whereas treatments C0 and C1 for the same trait did).
Table 6 reports the accuracy measures (adjusted R
2, RMSE, and CV
RMSE) of all linear models constructed considering all 39 possible inputs (before input selection), along with the number of predictors (input variables) selected by means of the lasso approach. At the tillering stage (BBCH 25), high correlations were only observed for BT
25 and NU
25. Thus, this time point appeared to be too soon for predicting the future development of the plants, particularly, when the first TdF had not been applied yet. Nevertheless, BT
25 and NU
25 were considered the two most important traits to support N fertilization dose (units/ha) at tillering. The linear models constructed for the booting stage using two UAV images (at BBCH 25 and 45) exhibited high correlations for most of the agronomic traits. The models for biomass (BT
99 and BSL
99) and N concentration (NC
99, NCSL
99, and NUG) at maturity achieved relatively lower accuracies, but in most cases the adjusted R
2 values were higher than 0.7, with exception to HI. The linear models constructed for the booting stage, using three UAV images (at BBCH 25, 31, and 45), generally resulted in increased accuracies compared to the corresponding models constructed with only two images, although this was not always the case. The models that did exhibit an increase in accuracy always comprised a higher number of predictors than those with two images, which means that they exploited the additional information provided by the image at BBCH 31, as intuitively expected. Notable exceptions were the models for NUSL
99 and NUG, which resulted in higher accuracy with two images, but with overly complex models. In this case, the additional information provided by the image at BBCH 31 enabled the simplification of the models, but with a penalty in accuracy at the same time. It is worth noting that for most of the agronomic traits (PH
45, BT
45, yield, NC
45, and all NU traits), the models with only two images could achieve competitive performance. In all cases, the number of predictors selected by lasso was low, especially if we took into consideration the very high number of available predictors (see
Section 2.3). Specifically, the average number of predictors for all models was 4.39, with a median value of 4. For the models constructed for the tillering stage, 10.83% of the available predictors were used on average by the models, 5.77% for the models at booting with two images, and 3.85% for those at booting with three images. As such, the models could be easily visualized and the most important input variables could be identified, whereas the computational and—perhaps most importantly—storage requirements remained relatively low.
In order to give a visual representation of the models’ errors,
Figure 3 depicts the scatter plots of predicted versus observed values for some indicative models constructed in each stage. The gray dashed line represents the ideal perfect linear relationship, whereas the continuous red line is the simple linear regression line of the predicted versus observed data. Models with high adjusted R
2 values exhibited, generally, uniform distribution of data around the ideal prediction line, with the regression line being very close to the latter and very few outliers observed in a few cases. For completeness, the scatter plots for all models constructed are provided as
Supplementary Materials.
Table 7 presents the linear models constructed at the tillering stage, whereas
Table 8 and
Table 9 present the models constructed at the booting stage using features extracted from two and three UAV images, respectively. For convenience, the adjusted R
2 and RMSE values of
Table 6 are replicated in these tables as well. Focusing on the models constructed at the tillering stage that exhibit high accuracy (BT
25, NC
99, and NU
25 in
Table 7), they mostly exploited chlorophyll-sensitive vegetation indices (gRDVI, CI
G, CI
RE, TCARI, MCARI, MRETVI) together with the green and red reflectance channels. At the BBCH 25 stage, the canopy was not fully closed, introducing strong background soil/water effects. This in turn decreased the efficiency of traditional biomass-sensitive VIs (e.g., NDVI, SR, and mSR) for estimating agronomic traits at the tillering stage. Conversely, chlorophyll-sensitive VIs incorporated the green or RE channel (or both), which rendered them more sensitive to small variations of biomass. The latter was also true for the model built for yield, although the latter exhibited relatively lower accuracy (Adj. R
2 = 0.61; RMSE = 1.47 tn∙ha
−1) than the aforementioned ones.
Regarding the models built at the booting stage (
Table 8 and
Table 9), models for biomass-related traits (height and biomass) tended to exploit traditional biomass-sensitive VIs such as NDVI, SR, mSR, or NDRE. Indeed, those VIs were found to exhibit high correlations with biomass in a number of vegetation monitoring studies. At the booting stage, the canopy was fully closed and those VIs could provide the necessary information for the differences in biomass, since no significant background effects were observed. The models created for yield also comprised biomass-sensitive VIs, together with the red and/or green channels. Conversely, the models built for N-related traits (NU and NC) comprised mostly VIs sensitive to chlorophyll (e.g., MTCARI, MCARI2, and TCARI/OSAVI) and VIs incorporating the green channel (e.g., CI
G and gNDVI), along with the traditional biomass-sensitive VIs in some cases. Plant N content was highly correlated with its chlorophyll content and, hence, chlorophyll-sensitive VIs increased the predictive capabilities of models for N-related traits. Moreover, the recently developed VIs [
32], incorporating the RE together with the green channel, were frequently selected (most notably MRETVI and REGNDVI) for all model categories. Finally, the original green and red channels (reflectance values) were also frequently selected at both stages, which shows that the original image could increase the model’s accuracy.
Since the agronomic traits’ estimation was performed using inputs derived from UAV imagery, the models’ output or errors could be displayed on the spatial domain, in order to support decision making.
Figure 4 provides such an example, depicting each experimental plot’s yield prediction errors in 2017, for the models constructed at the tillering and the booting stage. Error values were calculated as predicted minus field-measured yield (tn∙ha
−1). For most plots, the errors greatly decreased (in absolute terms) from the tillering to the booting stage, as expected from the numerical results presented so far. Using three images for the models built at the booting stage further decreased the absolute errors, that is, more plots exhibited error values closer to zero. A notable exception was the top row’s fifth plot from the left, for which the absolute error increased significantly, but this was a rather isolated case. Similar maps of the models’ outputs can directly support decision making, in order to efficiently adapt the applied treatment during the growing season.
Finally,
Table 10 reports the accuracy measures of all linear models constructed considering the 23 possible inputs (before input selection) that do not depend on the RE channel, in a similar format with
Table 6 above. For some models, the accuracy measures’ values equaled those of
Table 6, which was observed when the original models did not include any RE-related input. For most of the rest, their accuracy was inferior, but the difference was usually small (relative decrease in Adj. R
2 below 10%). This indicates that multispectral sensors without a RE channel can be used for employing the proposed methodology, with perhaps a small penalty in predictive accuracy. In a few cases, the accuracy achieved by the non-RE-dependent model was even higher than the RE-dependent ones, which is a result of the higher number of inputs selected by the lasso process for the former models. This is possible since no input selection methodology is perfect or, more precisely, lasso solves the minimization problem of equation (1), but that does not guarantee the maximization of the linear model’s accuracy built independently in a subsequent step. Nevertheless, this behavior was observed rather rarely, at least for the models that exhibit high correlations with the observed data (e.g., Adj. R
2 greater than 0.7). For brevity in the presentation here, the non-RE-dependent models’ formulas are provided as
Supplementary Materials.
4. Discussion
Estimating N requirements and biomass-related traits in rice plants is crucial for adopting efficient precision agriculture management practices. The analysis conducted showcased that it is possible to estimate at least BT and NU at key stages within the growing season (that is, a few days before the two TdFs) using UAV-collected multispectral images and derivative VIs. At the booting stage, estimating PH, yield, and NC was also achieved with low fractions of variance unexplained. However, estimating NC before the canopy closes when the plant biomass is less compact (i.e., at tillering) is a great challenge, as observed in a number of previous studies [
22,
31,
32,
65]. Plant biomass dominates canopy reflectance before the heading stage, making the estimation of chlorophyll and N concentration at early growth stages difficult [
65]. Nevertheless, an R
2 value of 80% was achieved for NU by the model built at tillering, which is an encouraging result. Due to the contribution of biomass and canopy structure before the heading stage and given that NU is the combination of biomass and NC, NU is more closely associated with VIs at the early growth stages [
22].
Conversely, no acceptable accuracy was able to be achieved for HI at any growth stage. The improvement in HI is a consequence of increased grain population density coupled with stable individual grain weight, both of which are genotypic. Its high heritability was showcased by examining its rather weak response to variation in management practices (fertilization, population density, application of growth regulators) in the absence of severe stress [
101]. Management practices that have been proposed for increasing HI of rice are based on special water treatments [
102], none of which was applied in our experimentation setups. As such, no significant variations in HI across different N treatments was observed in the field data, as shown in
Figure 2l.
Our modeling approach was based on multivariate linear regression exploiting several biomass- and chlorophyll-sensitive VIs, since the use of reflectance in individual bands alone has limitations due to the overlap of the effect of different nutrients and the influence of leaf and structural canopy parameters [
103]. The linear regression approach in combination with lasso was selected so that the derived models are interpretable. The analysis showed that chlorophyll-sensitive VIs comprising the green and/or the RE bands are required for estimating N content at all stages and biomass at the early growth stages, whereas traditional biomass-sensitive VIs are useful for estimating biomass-relevant traits after canopy closure, which is in line with previous studies (e.g., [
32,
34,
37]). Moreover, the reflectance values of the camera green and red bands were selected for most models at both the tillering and booting stages.
The modeling approach design tried to minimize the image acquisitions during the growing season. As such, only two images were required for estimating BT and NU a few days before each of the two TdFs. Yield prediction accuracy was higher at the booting stage (R
2 from 0.61 at tillering to 0.77 at booting with two images), which is consistent with a recent study [
39] that showed that the best timing for predicting yield from hyperspectral measurements was indeed the booting stage. The alternative approach of using an intermediate image approximately seven to ten days after the first TdF is useful only in certain cases and, especially, if one is required to predict the plant’s N content (NU or NC) at the maturity stage. Yield prediction accuracy is also slightly increased (R
2 from 0.77 to 0.80) with the use of the additional image data. Nevertheless, two UAV images will be required in most real-world scenarios. It should be noted that the image acquisition stages reported here (BBCH 25, 31, and 45) are mostly indicative; the results are not expected to change if images at plus/minus one value of the BBCH scale are used instead, since the plant’s physiology changes marginally in a two–3-day period of time.
The results presented in this study can be very useful for providing N recommendations for the two TdFs in rice. One possibility is to employ the approach of Xue et al. [
28], which uses the plant’s NU at each stage and the soil N supply, which can be estimated indirectly from a treatment with no N applied (C0 in our experimentation). Taking into consideration that the soil initial N supply is mandatory, since the amount of N not accounted for N recovery efficiency should not be interpreted as N that is lost without utilization from the soil–plant system. It has been reported that the recovery efficiency of N fertilizer by rice generally ranges from 20% to 80% with an average of about 30% to 40%, whereas 16% to 25% of the total applied N fertilizer could be recovered in the soil organic fraction between panicle differentiation and maturity of rice [
104]. An alternative approach is to use the models’ estimations of biomass and NU or NC to calculate the plant’s N nutrition index (NNI), provided that a critical N dilution curve for the region of application and selected rice variety exists or can be calibrated [
34,
36,
65]. A third alternative is to use the N concentration estimations derived from the models to calculate the so-called nitrogen sufficiency index (NSI), which can be directly used to provide N fertilization recommendations [
105]. This approach requires the establishment of reference strips within the rice field, which receive an amount of N equal or slightly higher than that recommended by standard soil tests [
106].
No matter which of the aforementioned approaches is selected for providing fertilization recommendations, the methodology presented in this study provides a cost-effective alternative to sampling approaches (either in-situ SPAD measurements or N concentration or uptake calculated in the laboratory) for estimating the rice plant’s N status. Moreover, the use of UAV-collected spatial data highlights the within-field variability of N requirements directly, since the agronomic traits estimations from the models are produced for all image pixels, thus resulting in a detail map of the whole field. The traditional approach of field sampling requires spatial interpolation between the sampled positions for obtaining equivalent maps, which generally introduces large errors in the estimations unless an unrealistically dense sampling is performed. It should be stressed that our approach does not provide any insights on the most appropriate timing to apply the TdFs. In this study we have followed the local and international standard practices for rice cultivation, with the two TdFs being performed at tillering and at booting according to the international standards. If more precision with respect to the optimal timing for applying the TdFs is required, growth simulation models that are based on meteorological predictions [
107], time-series of remotely sensed data [
62], or both [
108] can be used.
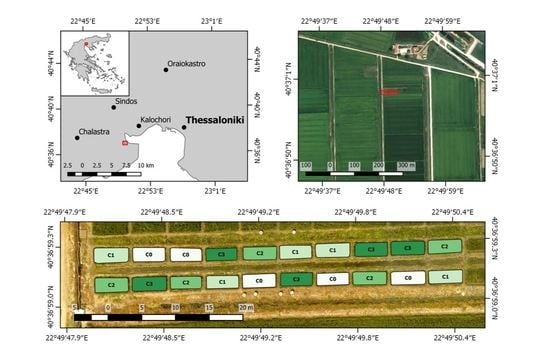
The models presented in this study were constructed considering small experimental plots, in order to facilitate field sampling and to assure that each sampling area is homogeneous, at least as much as possible. For real-world field-level applications, one has to also address the issue of accurate geo-referencing of the orthomosaic derived from the original UAV imagery. The sensitivity of global navigation satellite system (GNSS) receivers and inertial measurement units (IMU) integrated within compact sensors, such as Parrot Sequoia, will typically result in geolocation errors of 3–5 m in the orthorectified mosaic produced by most SfM software, which may be unacceptably high for precision agriculture applications, especially in smaller rice paddies. This issue can be addressed through the use of ground control points (GCPs) during image acquisition [
83,
109], the location of which is either fixed or measured with specialized equipment more accurate than the camera integrated one (e.g., differential GNSS). Since the latter could prove quite cumbersome and time-consuming for large-scale applications (especially in rice fields which are flooded), automatic registration methods using a reference image can be applied instead, which is an active research topic with many methods having been proposed recently [
110,
111,
112,
113]. The latter can reduce the geolocation errors to approximately 1 m or even less, which is sufficient for precision agriculture applications and variable rate fertilization in particular. From our experience, the Pix4D Mapper Pro software—that fuses SfM techniques with Sequoia’s GNSS and IMU readings during the flight and known camera/image acquisition parameters—was able to produce accurate orthomosaics in relative terms, that is, the distances between points were accurate but the whole orthomosaic exhibited shifts of 1–3 m and a very slight rotation in some cases. The subject of accurate UAV image registration is very broad and outside the scope of the current study. To this end, we actually shifted the polygons used to derive the average pixels value within each plot manually for each image, so that they corresponded to the same locations in all UAV image mosaics. Differences of a few centimeters that may have been introduced by this process are insignificant for our analysis, since the experimental plots are considered homogeneous.
For large-scale applications (big farms with scattered fields or regional level), high-resolution satellite imagery could be considered for applying our models, if UAV image acquisition is deemed to be too time or resource inefficient. For large rice fields, Sentinel-2 imagery is an obvious candidate, since it comprises a RE channel and has a short revisit cycle (five days). However, the level of analysis would be much coarser than ours, since the RE channel is provided at a spatial resolution of 20 m. Alternatively, only the 10 m bands of Sentinel-2 could be used in the analysis, employing the models built with only the 23 inputs not dependent on the RE channel (
Table 10 and
Tables S1–S3 in the Supplementary Materials). Commercial, very high-resolution satellite imagery is another viable alternative, at least within the framework of a commercial fertilization recommendation service provision. The models constructed with only the VIs that do not require the existence of a RE channel are useful in such a scenario, since most commercial, very high-resolution satellite sensors do not comprise an RE channel. In the latter theme, it is worth mentioning the products that have started to be offered recently by Planet (
https://www.planet.com), a company that operates a large constellation of small satellites (Cubesats) that acquire daily images of the whole earth. Both PlanetScope (spatial resolution of 3 m; images acquired daily) and SkySat (spatial resolution of 0.8–1 m; sub-daily acquisitions are possible) analysis-ready data could prove useful for establishing a fertilization recommendation service based on the proposed modeling approach, since their spatial and temporal resolution is sufficient for obtaining cloud-free imagery at the required growth stages for every rice field in a broader region.
Perhaps the most important novelty of our study is the use of a cost-effective multispectral imaging sensor onboard UAVs for estimating rice agronomic traits. Most other studies for estimating rice agronomic traits employ near-ground measurements, either using hyperspectral sensors or active canopy ones, which however do not need to account for the complex background phenomena arising for the lower resolution of the UAV-collected imagery. In the latter case, techniques for removing the background [
57] are difficult to be employed. This also means that the results of previous studies are not necessarily transferable to UAV-collected multispectral data. In this study we showcase that some similarities with previous results do exist, at least in terms of the most important VIs for estimating rice agronomic traits. Arguably, our study provides a more applied approach that could be integrated to any workflow or budget and can be easily adapted to a wide range of applications, even commercial ones, at least with the current technological standards.
The predictive models presented in this study were developed following field experimentation in Greece using a japonica-type rice variety. Rice in Europe is mainly cultivated in the South Mediterranean countries, which are characterized by very similar climatic conditions, whereas the cultivation treatments are also similar. Provided that no extreme variations in meteorological conditions are observed during the growing season, the results of this study should be readily applicable to other rice producing European countries, although this needs to be testified with additional experimentation in the future. Moreover, the sensor used (Parrot® Sequoia™) was selected because it provides a generally affordable solution that is readily supported by most flight planning and orthomosaic generation software, without the need of any additional configuration on the user side. However, its bands’ spectral response is very similar with that of other alternatives or even very-high resolution satellite imagers. Therefore, our workflow could be applied considering a number of alternative sensors.