Estimation of Paddy Rice Nitrogen Content and Accumulation Both at Leaf and Plant Levels from UAV Hyperspectral Imagery
Abstract
:1. Introduction
2. Materials and Methods
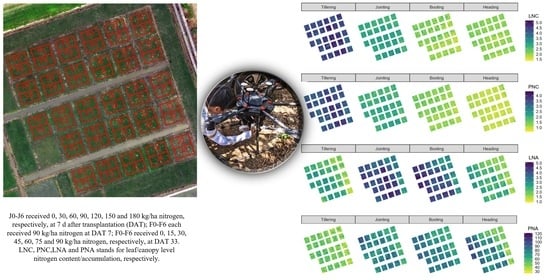
2.1. Study Area and Experimental Setup
2.2. Field Data Collection
2.3. Methods
| Algorithm 1: Hyperparameter optimization and model calibration algorithm. |
 |
3. Results
3.1. Variability in the Measured Nitrogen, Chlorophyll and Aboveground Biomass
3.2. Performance of the VIs
3.3. Performance of the PLSR and ML Methods
3.4. Evaluation on Hyperspectral Imageries
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Berger, K.; Verrelst, J.; Féret, J.B.; Wang, Z.; Wocher, M.; Strathmann, M.; Danner, M.; Mauser, W.; Hank, T. Crop nitrogen monitoring: Recent progress and principal developments in the context of imaging spectroscopy missions. Remote Sens. Environ. 2020, 242, 111758. [Google Scholar] [CrossRef]
- Kokaly, R.F.; Asner, G.P.; Ollinger, S.V.; Martin, M.E.; Wessman, C.A. Characterizing canopy biochemistry from imaging spectroscopy and its application to ecosystem studies. Remote Sens. Environ. 2009, 113, S78–S91. [Google Scholar] [CrossRef]
- Li, D.; Zhang, P.; Chen, T.; Qin, W. Recent Development and Challenges in Spectroscopy and Machine Vision Technologies for Crop Nitrogen Diagnosis: A Review. Remote Sens. 2020, 12, 2578. [Google Scholar] [CrossRef]
- Lemaire, G.; Jeuffroy, M.H.; Gastal, F. Diagnosis tool for plant and crop N status in vegetative stage. Eur. J. Agron. 2008, 28, 614–624. [Google Scholar] [CrossRef]
- Chlingaryan, A.; Sukkarieh, S.; Whelan, B. Machine learning approaches for crop yield prediction and nitrogen status estimation in precision agriculture: A review. Comput. Electron. Agric. 2018, 151, 61–69. [Google Scholar] [CrossRef]
- Onojeghuo, A.O.; Blackburn, G.A.; Huang, J.; Kindred, D.; Huang, W. Applications of satellite ‘hyper-sensing’ in Chinese agriculture: Challenges and opportunities. Int. J. Appl. Earth Obs. Geoinf. 2018, 64, 62–86. [Google Scholar] [CrossRef] [Green Version]
- Pancorbo, J.; Camino, C.; Alonso-Ayuso, M.; Raya-Sereno, M.; Gonzalez-Fernandez, I.; Gabriel, J.; Zarco-Tejada, P.; Quemada, M. Simultaneous assessment of nitrogen and water status in winter wheat using hyperspectral and thermal sensors. Eur. J. Agron. 2021, 127, 126287. [Google Scholar] [CrossRef]
- Huang, Z.; Turner, B.J.; Dury, S.J.; Wallis, I.R.; Foley, W.J. Estimating foliage nitrogen concentration from HYMAP data using continuum removal analysis. Remote Sens. Environ. 2004, 93, 18–29. [Google Scholar] [CrossRef]
- Yang, G.; Zhao, C.; Pu, R.; Feng, H.; Li, Z.; Li, H.; Sun, C. Leaf nitrogen spectral reflectance model of winter wheat ( Triticum aestivum ) based on PROSPECT: Simulation and inversion. J. Appl. Remote Sens. 2015, 9, 095976. [Google Scholar] [CrossRef]
- Berger, K.; Verrelst, J.; Féret, J.B.; Hank, T.; Wocher, M.; Mauser, W.; Camps-Valls, G. Retrieval of aboveground crop nitrogen content with a hybrid machine learning method. Int. J. Appl. Earth Obs. Geoinf. 2020, 92, 102174. [Google Scholar] [CrossRef]
- Jacquemoud, S.; Baret, F. PROSPECT: A model of leaf optical properties spectra. Remote Sens. Environ. 1990, 34, 75–91. [Google Scholar] [CrossRef]
- Baret, F.; Houles, V.; Guerif, M. Quantification of plant stress using remote sensing observations and crop models: The case of nitrogen management. J. Exp. Bot. 2006, 58, 869–880. [Google Scholar] [CrossRef] [Green Version]
- Tian, Y.; Yao, X.; Yang, J.; Cao, W.; Zhu, Y. Extracting Red Edge Position Parameters from Ground- and Space-Based Hyperspectral Data for Estimation of Canopy Leaf Nitrogen Concentration in Rice. Plant Prod. Sci. 2011, 14, 270–281. [Google Scholar] [CrossRef]
- Li, F.; Mistele, B.; Hu, Y.; Yue, X.; Yue, S.; Miao, Y.; Chen, X.; Cui, Z.; Meng, Q.; Schmidhalter, U. Remotely estimating aerial N status of phenologically differing winter wheat cultivars grown in contrasting climatic and geographic zones in China and Germany. Field Crop. Res. 2012, 138, 21–32. [Google Scholar] [CrossRef]
- Patel, M.K.; Ryu, D.; Western, A.W.; Suter, H.; Young, I.M. Which multispectral indices robustly measure canopy nitrogen across seasons: Lessons from an irrigated pasture crop. Comput. Electron. Agric. 2021, 182, 106000. [Google Scholar] [CrossRef]
- Li, F.; Miao, Y.; Hennig, S.D.; Gnyp, M.L.; Chen, X.; Jia, L.; Bareth, G. Evaluating hyperspectral vegetation indices for estimating nitrogen concentration of winter wheat at different growth stages. Precis. Agric. 2010, 11, 335–357. [Google Scholar] [CrossRef]
- Fitzgerald, G.; Rodriguez, D.; Leary, G.O. Measuring and predicting canopy nitrogen nutrition in wheat using a spectral index—The canopy chlorophyll content index ( CCCI ). Field Crops Res. 2010, 116, 318–324. [Google Scholar] [CrossRef]
- Lee, Y.J.; Yang, C.M.; Chang, K.W.; Shen, Y. A Simple Spectral Index Using Reflectance of 735 nm to Assess Nitrogen Status of Rice Canopy. Agron. J. 2008, 100, 205–212. [Google Scholar] [CrossRef]
- Stroppiana, D.; Boschetti, M.; Brivio, P.A.; Bocchi, S. Plant nitrogen concentration in paddy rice from field canopy hyperspectral radiometry. Field Crop. Res. 2009, 111, 119–129. [Google Scholar] [CrossRef]
- Tian, Y.C.; Yao, X.; Yang, J.; Cao, W.X.; Hannaway, D.B.; Zhu, Y. Assessing newly developed and published vegetation indices for estimating rice leaf nitrogen concentration with ground- and space-based hyperspectral reflectance. Field Crops Res. 2011, 120, 299–310. [Google Scholar] [CrossRef]
- Wang, L.; Chang, Q.; Li, F.; Yan, L.; Huang, Y.; Wang, Q.; Luo, L. Effects of Growth Stage Development on Paddy Rice Leaf Area Index Prediction Models. Remote Sens. 2019, 11, 361. [Google Scholar] [CrossRef] [Green Version]
- Moharana, S.; Dutta, S. Spatial variability of chlorophyll and nitrogen content of rice from hyperspectral imagery. ISPRS J. Photogramm. Remote Sens. 2016, 122, 17–29. [Google Scholar] [CrossRef]
- Verrelst, J.; Malenovský, Z.; Van der Tol, C.; Camps-Valls, G.; Gastellu-Etchegorry, J.P.; Lewis, P.; North, P.; Moreno, J. Quantifying Vegetation Biophysical Variables from Imaging Spectroscopy Data: A Review on Retrieval Methods. Surv. Geophys. 2019, 40, 589–629. [Google Scholar] [CrossRef] [Green Version]
- Hansen, P.M.; Schjoerring, J.K. Reflectance measurement of canopy biomass and nitrogen status in wheat crops using normalized difference vegetation indices and partial least squares regression. Remote Sens. Environ. 2003, 86, 542–553. [Google Scholar] [CrossRef]
- Li, F.; Mistele, B.; Hu, Y.; Chen, X.; Schmidhalter, U. Reflectance estimation of canopy nitrogen content in winter wheat using optimised hyperspectral spectral indices and partial least squares regression. Eur. J. Agron. 2014, 52, 198–209. [Google Scholar] [CrossRef]
- Onoyama, H.; Ryu, C.; Suguri, M.; Iida, M. Nitrogen prediction model of rice plant at panicle initiation stage using ground-based hyperspectral imaging: Growing degree-days integrated model. Precis. Agric. 2015, 16, 558–570. [Google Scholar] [CrossRef]
- Yao, X.; Huang, Y.; Shang, G.; Zhou, C.; Cheng, T.; Tian, Y.; Cao, W.; Zhu, Y. Evaluation of six algorithms to monitor wheat leaf nitrogen concentration. Remote Sens. 2015, 7, 14939–14966. [Google Scholar] [CrossRef] [Green Version]
- Huang, S.; Miao, Y.; Yuan, F.; Gnyp, M.; Yao, Y.; Cao, Q.; Wang, H.; Lenz-Wiedemann, V.; Bareth, G. Potential of RapidEye and WorldView-2 Satellite Data for Improving Rice Nitrogen Status Monitoring at Different Growth Stages. Remote Sens. 2017, 9, 227. [Google Scholar] [CrossRef] [Green Version]
- Wang, B.; Chen, J.; Ju, W.; Qiu, F.; Zhang, Q.; Fang, M.; Chen, F. Limited Effects of Water Absorption on Reducing the Accuracy of Leaf Nitrogen Estimation. Remote Sens. 2017, 9, 291. [Google Scholar] [CrossRef] [Green Version]
- Li, F.; Wang, L.; Liu, J.; Wang, Y.; Chang, Q. Evaluation of Leaf N Concentration in Winter Wheat Based on Discrete Wavelet Transform Analysis. Remote Sens. 2019, 11, 1331. [Google Scholar] [CrossRef] [Green Version]
- Marang, I.J.; Filippi, P.; Weaver, T.B.; Evans, B.J.; Whelan, B.M.; Bishop, T.F.A.; Murad, M.O.F.; Al-Shammari, D.; Roth, G. Machine Learning Optimised Hyperspectral Remote Sensing Retrieves Cotton Nitrogen Status. Remote Sens. 2021, 13, 1428. [Google Scholar] [CrossRef]
- Nigon, T.; Yang, C.; Dias Paiao, G.; Mulla, D.; Knight, J.; Fernández, F. Prediction of Early Season Nitrogen Uptake in Maize Using High-Resolution Aerial Hyperspectral Imagery. Remote Sens. 2020, 12, 1234. [Google Scholar] [CrossRef] [Green Version]
- Zheng, H.; Li, W.; Jiang, J.; Liu, Y.; Cheng, T.; Tian, Y.; Zhu, Y.; Cao, W.; Zhang, Y.; Yao, X. A Comparative Assessment of Different Modeling Algorithms for Estimating Leaf Nitrogen Content in Winter Wheat Using Multispectral Images from an Unmanned Aerial Vehicle. Remote Sens. 2018, 10, 2026. [Google Scholar] [CrossRef] [Green Version]
- Yi, Q.; Huang, J.; Wang, F.; Wang, X. Evaluating the performance of PC-ANN for the estimation of rice nitrogen concentration from canopy hyperspectral reflectance. Int. J. Remote Sens. 2010, 31, 931–940. [Google Scholar] [CrossRef]
- Wang, F.; Huang, J.; Wang, Y.; Liu, Z.; Zhang, F. Estimating nitrogen concentration in rape from hyperspectral data at canopy level using support vector machines. Precis. Agric. 2013, 14, 172–183. [Google Scholar] [CrossRef]
- Guo, J.; Zhang, J.; Xiong, S.; Zhang, Z.; Wei, Q.; Zhang, W.; Feng, W.; Ma, X. Hyperspectral assessment of leaf nitrogen accumulation for winter wheat using different regression modeling. Precis. Agric. 2021, 1–25. [Google Scholar] [CrossRef]
- Wang, L.; Zhou, X.; Zhu, X.; Guo, W. Estimation of leaf nitrogen concentration in wheat using the MK-SVR algorithm and satellite remote sensing data. Comput. Electron. Agric. 2017, 140, 327–337. [Google Scholar] [CrossRef]
- Sun, J.; Yang, J.; Shi, S.; Chen, B.; Du, L.; Gong, W.; Song, S. Estimating Rice Leaf Nitrogen Concentration: Influence of Regression Algorithms Based on Passive and Active Leaf Reflectance. Remote Sens. 2017, 9, 951. [Google Scholar] [CrossRef] [Green Version]
- Dunn, B.; Dehaan, R.; Schmidtke, L.; Dunn, T.; Meder, R. Using Field-Derived Hyperspectral Reflectance Measurement to Identify the Essential Wavelengths for Predicting Nitrogen Uptake of Rice at Panicle Initiation. J. Infrared Spectrosc. 2016, 24, 473–483. [Google Scholar] [CrossRef]
- Zhao, H.; Song, X.; Yang, G.; Li, Z.; Zhang, D.; Feng, H. Monitoring of Nitrogen and Grain Protein Content in Winter Wheat Based on Sentinel-2A Data. Remote Sens. 2019, 11, 1724. [Google Scholar] [CrossRef] [Green Version]
- Moldenhauer, K.; Slaton, N. Rice growth and development. Rice Prod. Handb. 2001, 192, 7–14. [Google Scholar]
- Cerovic, Z.G.; Masdoumier, G.; Ghozlen, N.B.; Latouche, G. A new optical leaf-clip meter for simultaneous non-destructive assessment of leaf chlorophyll and epidermal flavonoids. Physiol. Plant. 2012, 146, 251–260. [Google Scholar] [CrossRef] [PubMed]
- Awais, M.; Li, W.; Cheema, M.J.M.; Hussain, S.; AlGarni, T.S.; Liu, C.; Ali, A. Remotely sensed identification of canopy characteristics using UAV-based imagery under unstable environmental conditions. Environ. Technol. Innov. 2021, 22, 101465. [Google Scholar] [CrossRef]
- Li, M.; Shamshiri, R.R.; Schirrmann, M.; Weltzien, C. Impact of Camera Viewing Angle for Estimating Leaf Parameters of Wheat Plants from 3D Point Clouds. Agriculture 2021, 11, 563. [Google Scholar] [CrossRef]
- Tian, M.; Ban, S.; Chang, Q.; Ma, W.; Yin, Z.; Wang, L. Estimation of SPAD value of cotton leaf using hyperspectral images from UAV-based imaging spectroradiometer. Trans. Chin. Soc. Agric. Eng. 2016, 47, 285–293. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- Kuhn, M. plsmod: Model Wrappers for Projection Methods. R Package Version 0.1.1. 2020. Available online: https://github.com/tidymodels/plsmod (accessed on 1 January 2021).
- Venables, W.N.; Ripley, B.D. Modern Applied Statistics with S, 4th ed.; Springer: New York, NY, USA, 2002; ISBN 0-387-95457-0. [Google Scholar]
- Liaw, A.; Wiener, M. Classification and Regression by randomForest. R News 2002, 2, 18–22. [Google Scholar]
- Karatzoglou, A.; Smola, A.; Hornik, K.; Zeileis, A. kernlab—An S4 Package for Kernel Methods in R. J. Stat. Softw. 2004, 11, 1–20. [Google Scholar] [CrossRef] [Green Version]
- Clarke, T.R.; Moran, M.S.; Barnes, E.M.; Pinter, P.J.; Qi, J. Planar domain indices: A method for measuring a quality of a single component in two-component pixels. Int. Geosci. Remote Sens. Symp. 2001, 3, 1279–1281. [Google Scholar] [CrossRef]
- Tucker, C.J. Red and photographic infrared linear combinations for monitoring vegetation. Remote Sens. Environ. 1979, 8, 127–150. [Google Scholar] [CrossRef] [Green Version]
- Huete, A. A soil-adjusted vegetation index (SAVI). Remote Sens. Environ. 1988, 25, 295–309. [Google Scholar] [CrossRef]
- Rondeaux, G.; Steven, M.; Baret, F. Optimization of soil-adjusted vegetation indices. Remote Sens. Environ. 1996, 55, 95–107. [Google Scholar] [CrossRef]
- Qi, J.; Chehbouni, A.; Huete, A.R.; Kerr, Y.H.; Sorooshian, S. A modified soil adjusted vegetation index. Remote Sens. Environ. 1994, 48, 119–126. [Google Scholar] [CrossRef]
- Gitelson, A.a.; Gritz, Y.; Merzlyak, M.N. Relationships between leaf chlorophyll content and spectral reflectance and algorithms for non-destructive chlorophyll assessment in higher plant leaves. J. Plant Physiol. 2003, 160, 271–282. [Google Scholar] [CrossRef] [PubMed]
- Sims, D.A.; Gamon, J.A. Relationships between leaf pigment content and spectral reflectance across a wide range of species, leaf structures and developmental stages. Remote Sens. Environ. 2002, 81, 337–354. [Google Scholar] [CrossRef]
- Haboudane, D.; Miller, J.R.; Tremblay, N.; Zarco-Tejada, P.J.; Dextraze, L. Integrated narrow-band vegetation indices for prediction of crop chlorophyll content for application to precision agriculture. Remote Sens. Environ. 2002, 81, 416–426. [Google Scholar] [CrossRef]
- Wu, C.; Niu, Z.; Tang, Q.; Huang, W. Estimating chlorophyll content from hyperspectral vegetation indices: Modeling and validation. Agric. For. Meteorol. 2008, 148, 1230–1241. [Google Scholar] [CrossRef]
- Dash, J.; Curran, P.J. The MERIS terrestrial chlorophyll index. Int. J. Remote Sens. 2004, 25, 5403–5413. [Google Scholar] [CrossRef]
- Guyot, G.; Baret, F. Utilisation de la haute resolution spectrale pour suivre l’etat des couverts vegetaux. Spectr. Signat. Objects Remote Sens. 1988, 287, 279–286. [Google Scholar]
- Cho, M.A.; Skidmore, A.K. A new technique for extracting the red edge position from hyperspectral data: The linear extrapolation method. Remote Sens. Environ. 2006, 101, 181–193. [Google Scholar] [CrossRef]
- Miller, J.R.; Hare, E.W.; Wu, J. Quantitative characterization of the vegetation red edge reflectance 1. An inverted-Gaussian reflectance model. Int. J. Remote Sens. 1990, 11, 1755–1773. [Google Scholar] [CrossRef]
- Yu, K.; Li, F.; Gnyp, M.L.; Miao, Y.; Bareth, G.; Chen, X. Remotely detecting canopy nitrogen concentration and uptake of paddy rice in the Northeast China Plain. ISPRS J. Photogramm. Remote Sens. 2013, 78, 102–115. [Google Scholar] [CrossRef]
- Schlemmera, M.; Gitelson, A.; Schepersa, J.; Fergusona, R.; Peng, Y.; Shanahana, J.; Rundquist, D. Remote estimation of nitrogen and chlorophyll contents in maize at leaf and canopy levels. Int. J. Appl. Earth Obs. Geoinf. 2013, 25, 47–54. [Google Scholar] [CrossRef] [Green Version]








| Index | Definition | Cite |
|---|---|---|
| NDVI | [52] | |
| SAVI | [53] | |
| OSAVI | [54] | |
| MSAVI | [55] | |
| CIgreen | [56] | |
| CIre | [56] | |
| NDRE | [17] | |
| D735 | [18] | |
| NDVI(483,503) | [19] | |
| TBVItian | [20] | |
| mND705 | [57] | |
| mSR705 | [57] | |
| TCARI | [58] | |
| TCARI/OSAVI | [58] | |
| TCARI2 | [59] | |
| OSAVI2 | [59] | |
| TCARI2/OSAVI2 | [59] | |
| MTCI | [60] | |
| REPLi | REP through linear four-point interpolation | [61] |
| REPLE | REP through linear extrapolation | [62] |
| mREIP | REP through Gaussian fit | [63] |
| CCCI | [17] | |
| NDPI | [14] |
| Growth Stage | Min | Max | Mean | CV | |
|---|---|---|---|---|---|
| LNC | Full Data set | 1.41 | 5.16 | 2.61 | 38.63 |
| Tillering | 4.00 | 5.16 | 4.65 | 7.69 | |
| Jointing | 2.43 | 3.30 | 2.84 | 8.93 | |
| Booting | 1.41 | 2.33 | 1.83 | 12.95 | |
| Heading | 1.77 | 2.32 | 2.07 | 6.52 | |
| PNC | Full Data set | 0.83 | 4.15 | 1.71 | 56.29 |
| Tillering | 3.09 | 4.15 | 3.68 | 8.91 | |
| Jointing | 1.70 | 2.37 | 2.00 | 9.87 | |
| Booting | 0.88 | 1.49 | 1.19 | 11.54 | |
| Heading | 0.83 | 1.20 | 1.01 | 9.90 | |
| LNA | Full Data set | 1.12 | 6.41 | 3.48 | 33.04 |
| Tillering | 1.26 | 2.93 | 2.05 | 24.21 | |
| Jointing | 2.89 | 6.41 | 4.29 | 22.48 | |
| Booting | 2.41 | 5.78 | 3.92 | 21.61 | |
| Heading | 1.12 | 4.23 | 2.64 | 27.31 | |
| PNA | Full Data set | 18.21 | 156.36 | 77.34 | 39.58 |
| Tillering | 18.21 | 39.00 | 28.01 | 21.83 | |
| Jointing | 46.75 | 95.54 | 65.96 | 19.91 | |
| Booting | 52.50 | 115.94 | 79.98 | 17.79 | |
| Heading | 55.61 | 141.83 | 95.72 | 20.64 | |
| LCC | Full Data set | 22.38 | 39.88 | 30.63 | 12.44 |
| Tillering | 33.78 | 39.88 | 37.36 | 4.55 | |
| Jointing | 28.67 | 35.46 | 31.57 | 5.09 | |
| Booting | 25.20 | 34.92 | 28.55 | 7.38 | |
| Heading | 27.69 | 34.37 | 30.26 | 5.08 | |
| CCC | Full Data set | 29.36 | 293.72 | 118.45 | 51.89 |
| Tillering | 32.71 | 81.63 | 56.20 | 23.99 | |
| Jointing | 65.51 | 201.15 | 122.81 | 27.85 | |
| Booting | 102.74 | 293.72 | 186.85 | 23.35 | |
| Heading | 29.36 | 127.69 | 86.03 | 26.14 | |
| ABG | Full Data set | 0.54 | 12.37 | 6.10 | 54.24 |
| Tillering | 0.54 | 0.98 | 0.75 | 14.86 | |
| Jointing | 2.56 | 4.19 | 3.28 | 13.54 | |
| Booting | 5.55 | 8.56 | 6.69 | 9.85 | |
| Heading | 6.69 | 11.99 | 9.36 | 13.43 |
| Calibration | Validation | |||||||
|---|---|---|---|---|---|---|---|---|
| Data Set | Vegetation Index | MREcv | RMSEcv | R | MREval | RMSEval | R | |
| LNC | Full Data set | TCARI/OSAVI | 25.85 | 0.74 | 0.62 | 24.95 | 0.68 | 0.61 |
| Tillering | CIgreen | 3.34 | 0.17 | 1.00 | 2.03 | 0.11 | 0.88 | |
| Jointing | REPLi | 3.80 | 0.12 | 1.00 | 4.06 | 0.13 | 0.87 | |
| Booting | CIre | 10.00 | 0.20 | 0.50 | 10.48 | 0.21 | 0.08 | |
| Heading | TCARI/OSAVI | 3.87 | 0.09 | 0.94 | 4.31 | 0.14 | 0.07 | |
| PNC | Full Data set | TCARI/OSAVI | 30.85 | 0.62 | 0.69 | 25.18 | 0.53 | 0.73 |
| Tillering | CIgreen | 3.45 | 0.14 | 1.00 | 2.25 | 0.10 | 0.90 | |
| Jointing | mND705 | 3.20 | 0.08 | 1.00 | 2.50 | 0.06 | 0.95 | |
| Booting | NDPI | 6.06 | 0.09 | 0.65 | 5.09 | 0.08 | 0.50 | |
| Heading | mND705 | 4.38 | 0.05 | 1.00 | 6.86 | 0.08 | 0.39 | |
| LNA | Full Data set | TBVITian | 16.91 | 0.61 | 0.72 | 17.82 | 0.74 | 0.69 |
| Tillering | D735 | 8.13 | 0.18 | 1.00 | 7.05 | 0.20 | 0.82 | |
| Jointing | OSAVI2 | 11.76 | 0.56 | 0.99 | 9.78 | 0.47 | 0.81 | |
| Booting | NDPI | 14.10 | 0.63 | 0.61 | 14.49 | 0.63 | 0.21 | |
| Heading | NDVI | 16.24 | 0.52 | 0.99 | 15.01 | 0.45 | 0.28 | |
| PNA | Full Data set | TCARI/OSAVI | 12.43 | 10.69 | 0.84 | 16.75 | 12.13 | 0.80 |
| Tillering | D735 | 7.52 | 2.26 | 1.00 | 6.78 | 2.27 | 0.85 | |
| Jointing | NDRE | 10.38 | 7.82 | 1.00 | 9.98 | 6.70 | 0.79 | |
| Booting | TCARI2/OSAVI2 | 8.19 | 7.75 | 0.74 | 9.53 | 10.75 | 0.36 | |
| Heading | REPLE | 10.30 | 11.44 | 1.00 | 13.35 | 13.94 | 0.51 | |
| Calibration (Cross-Validated) | Validation | |||||||
|---|---|---|---|---|---|---|---|---|
| Data Set | Method | MREcv | RMSEcv | MREval | RMSEval | |||
| LNC | Full Data set | ANN | 7.82 | 0.21 | 0.95 | 6.62 | 0.19 | 0.97 |
| Tillering | RF | 3.10 | 0.16 | 0.98 | 3.38 | 0.19 | 0.86 | |
| Jointing | SVM | 3.86 | 0.12 | 1.00 | 6.80 | 0.20 | 0.44 | |
| Booting | PLSR | 11.42 | 0.23 | 0.15 | 8.76 | 0.19 | 0.50 | |
| Heading | SVM | 3.66 | 0.09 | 0.94 | 10.54 | 0.25 | 0.01 | |
| PNC | Full Data set | ANN | 6.41 | 0.12 | 0.98 | 5.15 | 0.11 | 0.99 |
| Tillering | PLSR | 3.02 | 0.12 | 0.98 | 2.56 | 0.12 | 0.90 | |
| Jointing | SVM | 3.03 | 0.07 | 1.00 | 4.65 | 0.11 | 0.80 | |
| Booting | PLSR | 7.30 | 0.10 | 0.44 | 3.84 | 0.06 | 0.82 | |
| Heading | PLSR | 5.16 | 0.06 | 0.95 | 6.59 | 0.07 | 0.83 | |
| LNA | Full Data set | SVM | 14.85 | 0.60 | 0.72 | 11.04 | 0.49 | 0.81 |
| Tillering | ANN | 7.10 | 0.15 | 0.98 | 9.18 | 0.20 | 0.91 | |
| Jointing | PLSR | 11.09 | 0.53 | 0.98 | 10.88 | 0.45 | 0.84 | |
| Booting | ANN | 14.59 | 0.67 | 0.54 | 15.73 | 0.83 | 0.22 | |
| Heading | PLSR | 14.65 | 0.48 | 0.99 | 28.26 | 0.55 | 0.88 | |
| PNA | Full Data set | SVM | 11.32 | 9.16 | 0.84 | 11.11 | 11.40 | 0.87 |
| Tillering | PLSR | 6.02 | 1.79 | 1.00 | 9.31 | 2.80 | 0.95 | |
| Jointing | RF | 9.79 | 6.88 | 0.99 | 9.32 | 7.54 | 0.69 | |
| Booting | PLSR | 8.59 | 8.85 | 0.61 | 6.10 | 7.50 | 0.77 | |
| Heading | PLSR | 10.71 | 11.66 | 0.90 | 10.83 | 9.49 | 0.95 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, L.; Chen, S.; Li, D.; Wang, C.; Jiang, H.; Zheng, Q.; Peng, Z. Estimation of Paddy Rice Nitrogen Content and Accumulation Both at Leaf and Plant Levels from UAV Hyperspectral Imagery. Remote Sens. 2021, 13, 2956. https://0-doi-org.brum.beds.ac.uk/10.3390/rs13152956
Wang L, Chen S, Li D, Wang C, Jiang H, Zheng Q, Peng Z. Estimation of Paddy Rice Nitrogen Content and Accumulation Both at Leaf and Plant Levels from UAV Hyperspectral Imagery. Remote Sensing. 2021; 13(15):2956. https://0-doi-org.brum.beds.ac.uk/10.3390/rs13152956
Chicago/Turabian StyleWang, Li, Shuisen Chen, Dan Li, Chongyang Wang, Hao Jiang, Qiong Zheng, and Zhiping Peng. 2021. "Estimation of Paddy Rice Nitrogen Content and Accumulation Both at Leaf and Plant Levels from UAV Hyperspectral Imagery" Remote Sensing 13, no. 15: 2956. https://0-doi-org.brum.beds.ac.uk/10.3390/rs13152956